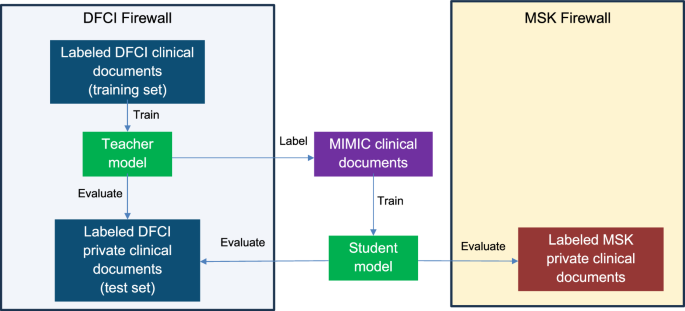
Assortment of information at DFCI and MSK was accredited by the Dana-Farber Harvard Cancer Heart Institutional Evaluation Board and the Memorial-Sloan Kettering Institutional Evaluation Board respectively.
Datasets
AACR Undertaking GENIE and GENIE BPC datasets
AACR Undertaking GENIE is a multi-institutional, worldwide consortium that collects deidentified tumor genomic subsequent technology sequencing information for evaluation and sharing2. GENIE has collected genomic information and fundamental scientific information, resembling cancer sort, affected person age at sequencing, and demographics, from effectively over 100,000 tumor specimens4. Memorial Sloan Kettering Cancer Heart (MSK) and Dana-Farber Cancer Institute (DFCI) are the 2 largest contributors to GENIE.
To extend the utility of the GENIE information for clinically related research questions by gathering information on granular longitudinal remedies and scientific outcomes, the AACR GENIE Biopharma Collaborative (BPC) venture was undertaken3. BPC includes annotation of EHRs to extract publicity variables, together with systemic remedy regimens, cancer histology, stage, and biomarkers; and outcomes, together with response, development, and metastatic websites31. MSK and DFCI are additionally the biggest contributors to GENIE BPC. In section I of BPC, records had been annotated for sufferers with non-small cell lung (NSCLC), colorectal, breast, pancreatic, prostate, or urothelial carcinoma. In section II, extra records for sufferers with NSCLC and colorectal cancer, plus records for sufferers with melanoma or renal cell, ovarian, or esophagogastric carcinoma, are being annotated. MSK and DFCI annotations out there as of November 2023 had been used for the present research, together with MSK sufferers with NSCLC, colorectal, breast, pancreatic, or prostate cancer; and DFCI sufferers with NSCLC, colorectal, breast, pancreatic, prostate, urothelial, or renal cell carcinoma. Annotation was carried out utilizing a Redcap database32. DFCI annotations had been cut up on the affected person stage into coaching (80%), validation/tuning (10%), and held-out take a look at (10%) units. MSK annotations had been used solely for mannequin analysis.
MIMIC-IV dataset
The Medical Data Mart for Intensive Care (MIMIC) dataset consists of deidentified structured and unstructured EHR information for sufferers who’ve been hospitalized within the intensive care unit at Beth Israel Deaconess Medical Heart. The present model of the dataset, MIMIC-IV, contains unstructured radiology reviews and discharge summaries for this cohort33. The MIMIC information can be found on request to credentialed researchers on Physionet34.
BPC information annotation
EHR information for sufferers included within the GENIE BPC cohorts, together with sufferers from each DFCI and MSK, had been manually annotated utilizing the Pathology, Radiology/Imaging, Indicators/Signs, Medical oncologist evaluation, and bioMarkers (PRISSMM) framework31,35. For the needs of the present research, this included annotation of every imaging report and one medical oncologist observe per 30 days alongside the illness trajectory. Imaging reviews had been annotated for a number of outcomes, together with ten derived variables: presence of cancer, development/worsening (“development”), response to remedy/enchancment (“response), and metastases to mind, bone, lung, liver, adrenal glands, lymph nodes, and peritoneum. Oncologist notes had been annotated for illness standing variables, together with three derived variables: presence of cancer, development, and response. This annotation strategy, and the utility of the curated outcomes as labels for NLP mannequin coaching at a single establishment, have been described beforehand5,6,36. PRISSMM-based outcomes, whether or not manually or AI-annotated, have additionally beforehand been proven to be related to total survival5,6,7.
NLP mannequin structure and coaching
First, multitask, Transformer37 -based neural community “trainer” fashions had been skilled to use the textual content of imaging reviews or oncologist notes for DFCI sufferers to predict the manually annotated outcomes described above.
For imaging reviews, a BERT-base-uncased21 mannequin obtained from Huggingface38 was fine-tuned in a multi-labeling setup with 10 binary classification heads to predict annotations of any cancer, development, response, and metastases to mind, bone, lung, liver, adrenal glands, lymph nodes, and peritoneum. Reviews had been truncated on the most BERT sequence size of 512 tokens. We name the ensuing mannequin, skilled instantly on DFCI imaging reviews, “DFCI-imaging-teacher.”
For medical oncologist notes, which regularly include extra tokens than the utmost of 512 that BERT fashions can deal with, a Medical-Longformer22,39 mannequin from Huggingface was fine-tuned with 3 binary classification heads to predict annotations of any cancer, development, and response. Oncologist notes had been left-truncated on the most Longformer sequence size of 4096 tokens. We name the ensuing mannequin, skilled on DFCI oncologist notes, “DFCI-medonc-teacher.”
For imaging reviews, coaching was carried out with a batch measurement of 16 on a single NVIDIA A6000 GPU. For oncologist notes, coaching was carried out with a batch measurement of 8 on a single NVIDIA A100 GPU with 80GB of VRAM. The AdamW optimizer was used40. Hyperparameters, together with studying fee, studying fee scheduler, and weight decay, had been tuned manually based mostly on efficiency within the validation/tuning set. Remaining fashions had been evaluated within the take a look at set. The loss perform utilized for every label was the binary cross entropy loss.
Subsequent, the DFCI-imaging-teacher and DFCI-medonc-teacher fashions had been used to label radiology reviews and discharge summaries, respectively, from the MIMIC-IV dataset. MIMIC radiology reviews had been restricted to paperwork addressing cancer or its absence by filtering for textual content together with the strings “cancer”, “restaging”, or “malignan*”, and to cancer-relevant imaging modalities by moreover requiring the strings “‘ct”, “mr”, “pet”, “nm”, or “mammo”. Equally, MIMIC discharge summaries had been restricted to paperwork addressing cancer or its absence by requiring the strings “cancer” or “malignan*”. DFCI-imaging-teacher inference was then run on the MIMIC radiology reviews, yielding the anticipated log odds of every of the ten imaging report labels. DFCI-medonc-teacher inference was run on the MIMIC discharge summaries, yielding predicted log odds of every of the three medical oncologist observe labels.
Lastly, “DFCI-imaging-student” and “DFCI-medonc-student” fashions had been skilled to use the textual content of the (deidentified) MIMIC radiology reviews and discharge summaries to predict the outputs (predicted log odds, or logits, of every label) of the DFCI-imaging-teacher and DFCI-medonc-teacher fashions respectively. The imply sq. error loss perform was used. This logit-matching setup is analogous to approaches generally used for mannequin distillation19. In distinction to the frequent distillation paradigm, nevertheless, our scholar fashions had the identical underlying structure because the trainer fashions however had been freshly skilled from BERT-base-uncased and ClinicalLongformer, respectively, so that they weren’t uncovered to PHI throughout coaching. Scholar mannequin hyperparameters had been manually tuned based mostly on efficiency amongst sufferers within the validation/tuning set of PHI-containing imaging reviews and oncologist notes from DFCI. Scholar mannequin efficiency was then evaluated within the held-out DFCI BPC PHI take a look at set. Lastly, scholar mannequin weights and inference code had been shared with MSK, which evaluated the fashions’ efficiency on their very own personal BPC-labeled paperwork.
We then in contrast our proposed distillation strategy, which includes a trainer mannequin skilled on labeled information, to merely utilizing a big language mannequin to hard-label a public dataset for scholar coaching. Given the resource-intensive nature of large-scale LLM inference, we took a random pattern of 20000 imaging reviews and discharge summaries from MIMIC and labeled them utilizing Llama-3-70B41. Llama was prompted to generate labels as described in Supplementary Notes 1 and 2. Scholar fashions (Llama-student-imaging and Llama-student-medonc) with the identical architectures as DFCI-imaging-student and DFCI-medonc-student had been then skilled on the MIMIC samples to predict the Llama-assigned labels. Lastly, to facilitate truthful comparability between the DFCI-teacher and Llama-teacher labeling technique with out confounding by coaching set measurement, we retrained variations of DFCI-imaging-student and DFCI-medonc-student on the identical samples of 20000 MIMIC paperwork every, yielding ‘DFCI-imaging-student-sample’ and ‘DFCI-medonc-student-sample’ fashions, respectively. Efficiency of those scholar fashions was then evaluated in the identical manner as the coed fashions skilled to predict labels assigned by the DFCI lecturers.
Mannequin coaching and inference had been carried out utilizing Pytorch, model 2.3.0.
A visible depiction of our total supervised teacher-student strategy is supplied in Fig. 1.
Subsequent, to illustrate the applicability of the teacher-student distillation strategy to duties past data extraction and classification, a BERT-based mannequin (DFCI-prognosis-teacher) was skilled to predict survival utilizing the textual content of particular person imaging reviews for sufferers within the DFCI coaching set. A adverse log-likelihood loss accommodating censored information was utilized. Efficiency was evaluated within the take a look at set utilizing the c-index. DFCI-prognosis-teacher was then used to label the MIMIC imaging reviews described above, and one other scholar (DFCI-prognosis-student) was skilled to predict these labels. Efficiency of DFCI-prognosis-student was then evaluated in each the DFCI and MSK take a look at units.
Mannequin analysis
Every human annotation-derived final result label was binary, and uncooked mannequin outputs corresponded to the anticipated log odds of every final result. The first mannequin final result metrics was the realm beneath the receiver working attribute curve (AUROC), the realm beneath the precision-recall curve (AUPRC), and the most effective F1 rating. The most effective F1 rating was outlined based mostly on the most effective F1 threshold within the dataset being evaluated. Analysis was carried out on a per-document foundation. Metrics had been in contrast to null values for every final result. For the AUROC, the null worth is 0.50. For the AUPRC, the null worth corresponds to p(final result), the prevalence of an final result within the analysis dataset. For the most effective F1 rating, the null worth is obtained by merely at all times guessing the optimistic class and corresponds to 2 × p(final result) / (p(final result) + 1).
To judge the vulnerability of the trainer and scholar fashions to a membership inference assault, a model of the oncologist observe trainer classification mannequin was skilled and overfitted to a pattern of 100 notes from the DFCI PHI coaching dataset. We then pulled a pattern of 100 notes from the DFCI PHI coaching/validation dataset. Inference utilizing the overfit trainer mannequin was carried out on all 200 notes. A easy logistic regression assault mannequin was then skilled on 60% of those 200 notes to predict whether or not a given observe was within the trainer’s coaching dataset. Inputs to the assault mannequin included the true guide labels (any cancer, development, and response), and the log odds of every of these outcomes generated by the trainer. The assault mannequin was then evaluated on the remaining 40% of the observe pattern utilizing the AUROC metric to seize its skill to discern whether or not notes had been used to prepare the trainer. The overfit trainer was used to re-label the MIMIC discharge abstract dataset. We then tried to prepare a model of the coed mannequin to predict the overfit trainer labels on these discharge summaries and prepare a brand new assault mannequin for the coed it in the identical manner.
Inclusion and ethics
Cohort eligibility was outlined as above, amongst sufferers who underwent tumor next-generation sequencing (NGS) for their cancers. At Dana-Farber, sufferers whose tumors underwent NGS on a research foundation supplied written knowledgeable consent; these whose tumors underwent NGS on a standard-of-care scientific foundation had been eligible based mostly on a waiver of knowledgeable consent given the minimal threat of the present research to contributors. At MSK, all sufferers supplied written knowledgeable consent. Race and ethnicity had been outlined based mostly on institutional medical document techniques, as reported by sufferers after they registered for scientific care. Sufferers weren’t compensated for participation.
Reporting abstract
Additional data on research design is out there within the Nature Portfolio Reporting Summary linked to this text.