Simply as ChatGPT generates textual content by predicting the phrase almost certainly to observe in a sequence, a brand new artificial intelligence (AI) model can write new proteins that aren’t naturally ocurring from scratch.
Scientists used the brand new model, ESM3, to create a brand new fluorescent protein that shares solely 58% of its sequence with naturally occurring fluorescent proteins, they mentioned in a examine revealed July 2 on the preprint bioRxiv database. Representatives from EvolutionaryScale, an organization fashioned by former Meta researchers, additionally outlined particulars June 25 in a statement.
The analysis crew has launched a small version of the model below a non-commercial license and can make the big model of the model obtainable to business researchers. In response to EvolutionaryScale, the know-how could possibly be helpful in fields starting from drug discovery to designing new chemical compounds for plastic degradation.
ESM3 is a big language model (LLM) just like OpenAI’s GPT-4, which powers the ChatGPT chatbot, and the scientists skilled their largest model on 2.78 billion proteins. For every protein, they extracted details about sequence (the order of the amino acid constructing blocks that make up the protein), construction (the three-dimensional folded form of the protein), and performance (what the protein does). They randomly masked items of details about these proteins and requested that ESM3 predict the lacking items.
They scaled this model up from analysis that the identical crew was conducting whereas nonetheless at Meta. In 2022 they announced EMSFold — a precursor to ESM3 that predicted unknown microbial protein constructions. That 12 months, Alphabet’s DeepMind additionally predicted protein structures for 200 million proteins.
Scientists subsequently identified that there are limitations to these AI models’ predictions and that the protein predictions have to be verified. However the strategies can nonetheless massively velocity up the search for protein constructions, as a result of the choice is to make use of X-rays to map out protein constructions one after the other — which is sluggish and expensive.
ESM3 goes past simply predicting current proteins, nevertheless. Utilizing the knowledge gleaned from 771 billion distinctive items of knowledge on construction, operate and sequence, the model can generate new proteins with explicit capabilities. It was described as a “ChatGPT moment for biology” by one of EvolutionaryScale’s backers.
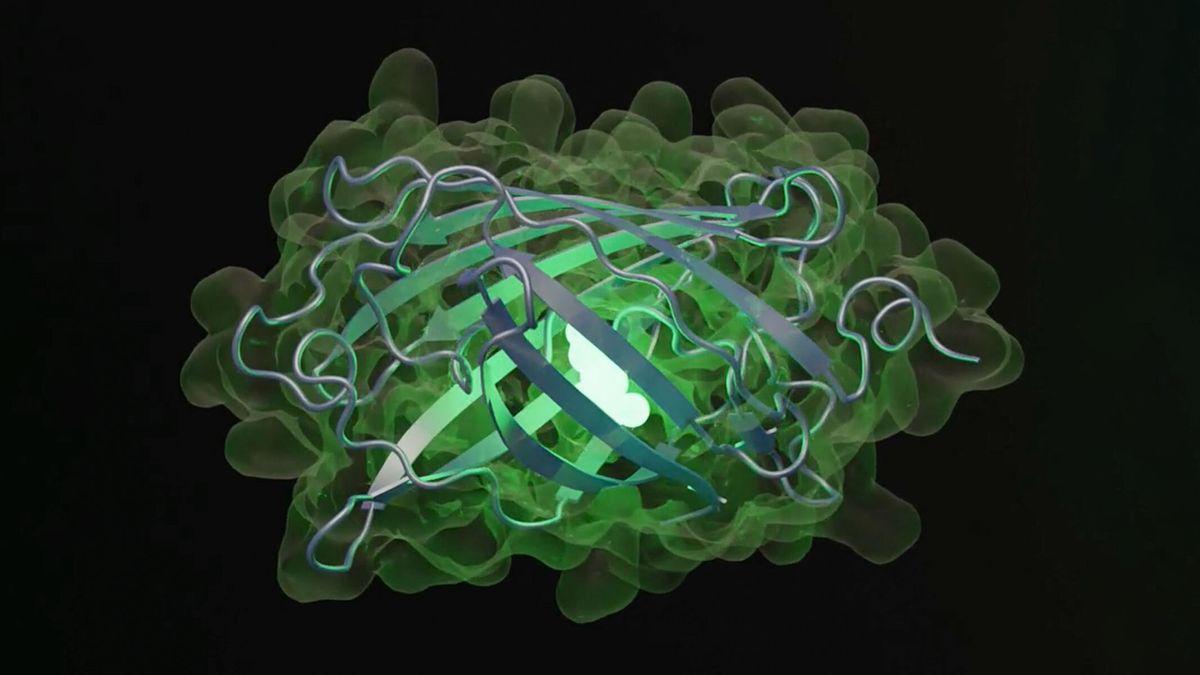
Within the new examine, the researchers queried the model to generate a brand new fluorescent protein — a sort of protein that captures mild and releases it again at an extended wavelength, making it shine in a brand new shade of inexperienced. These proteins are necessary for organic researchers who append them to molecules that they’re in learning to trace and picture them; their discovery and growth gained a Nobel Prize in chemistry in 2008.
The model generated 96 proteins with sequences and constructions prone to produce fluorescence. The researchers then selected one with the fewest sequences in widespread with naturally fluorescent proteins. Though this protein was 50 occasions much less vivid than pure inexperienced fluorescent proteins, ESM3 generated one other iteration that led to new sequences that elevated brightness — and the end result was a inexperienced fluorescent protein not like any found in nature, dubbed “esmGPF.” These iterations, performed in moments by the AI, would take 500 million years of evolution to attain, the EvolutionaryScale crew estimated.

“Proper now, we nonetheless lack the basic understanding of how proteins, particularly these “new to science,” behave when launched right into a residing system, however this can be a cool new step that permits us to strategy artificial biology in a brand new approach. AI modeling like ESM3 will allow the invention of latest proteins that the constraints of pure choice would by no means enable, creating improvements in protein engineering that evolution cannot. That’s thrilling. Nevertheless, the declare of simulating 500 million years of evolution focuses solely on particular person proteins, which doesn’t account for the numerous phases of pure choice that create the range of life we all know at this time. AI-driven protein engineering is intriguing, however I can’t assist feeling we could be overly assured in assuming we will outsmart the intricate processes honed by tens of millions of years of pure choice.”